
はじめに
この記事では、flash2を使った、ペアエンドリード(paired-end read)のマージ(read merge)、オーバーラップ(read overlap)とその他諸設定について紹介します。
flash2のもともとのアルゴリズムは、「FLASH (Fast Length Adjustment of SHort reads) 」と言われるものです。
名前からも推測できる通り、flash2は、FLASHの改良版となります。
FLASH は、リードの長さが2倍より短いDNA断片から生成されたペアエンドリードを、 正確かつ高速に、ペアエンドリードをマージする(リード同士を結合する)ためのツールの1つです。 マージされたリードペアは長いリードとして、リード数のカウントをしたり、 ゲノムアセンブリやゲノム解析のプロセスで活用されたりします。
このFLASHアルゴリズムでは、リードペア間の最小長以上のオーバーラップをすべて考慮し、ミスマッチ密度(オーバーラップ領域内のミスマッチ塩基の割合)が最も低くなるオーバーラップを選択しています。また、ミスマッチ部位の品質スコアも考慮しています。 マージされた配列を構築する際に、FLASHはオーバーラップした領域のコンセンサス配列を計算しています。
一方で、flash2の実行には、いくつかの制限があります。 当然ながら、オーバーラップしていないペアエンドリードのマージには使えません。 また、ペアエンドのfastqデータに適していて、サンガーシーケンスのデータには基本的に適していません。
FLASHの原著論文

Title: FLASH: fast length adjustment of short reads to improve genome assemblies Authors: Tanja Magoč and Steven L. Salzberg
詳しくは、原著論文を確認してみてください。
bioinformatics.oxfordjournals.org
今回のzsh実行環境
macOS Big Sur (バージョン11.5.2) MacBook Air (M1, 2020) チップ Apple M1 メモリ 16GB
M1 Macにおけるflash2のインストール
flash2は、GitHubからクローンして、ソースからコンパイルします。
コンパイルといっても、「ダウンロードとmake実行」でインストールが完了します。
おそらく最近のMacでも、ダウンロードには事前にgitコマンドを設定する必要があります。
gitコマンドの設定方法や使い方は、過去記事を参考のこと。
ターミナル(zsh)を起動して、以下のコマンドを実行していきます。
#ダウンロード git clone https://github.com/dstreett/FLASH2.git #ディレクトリ移動 cd FLASH2 #コンパイル make #パス設定の前段階: 手っ取り早く"/opt/homebrew/bin"に置いておく cd ../ mv FLASH2 /opt/homebrew/bin #HOMEに移動 #cd ~/ #パスを.zshrcを書き込む echo 'export PATH="/opt/homebrew/bin/FLASH2:$PATH"' >> .zshrc #いったんソースする source .zshrc #パス設定 which flash2 #/opt/homebrew/bin/FLASH2/flash2 #ヘルプで動作確認: 出力表示は補足に記載 flash2 -h
flash2によるペアエンドfastqのマージ実行
flash2の実行には、ペアエンドのfastqファイル(2つのfastq)が入力ファイルとして必要です。
GAGE (Genome Assembly Gold-standard Evaluation) というサイトがあって、 fastqのテストデータを色々と用意してくれているので、今回、それをテスト用のデータとして使うことにします。
詳しくは、GAGEの原著論文もあります。

GAGEからペアエンドのfastqデータを取得する
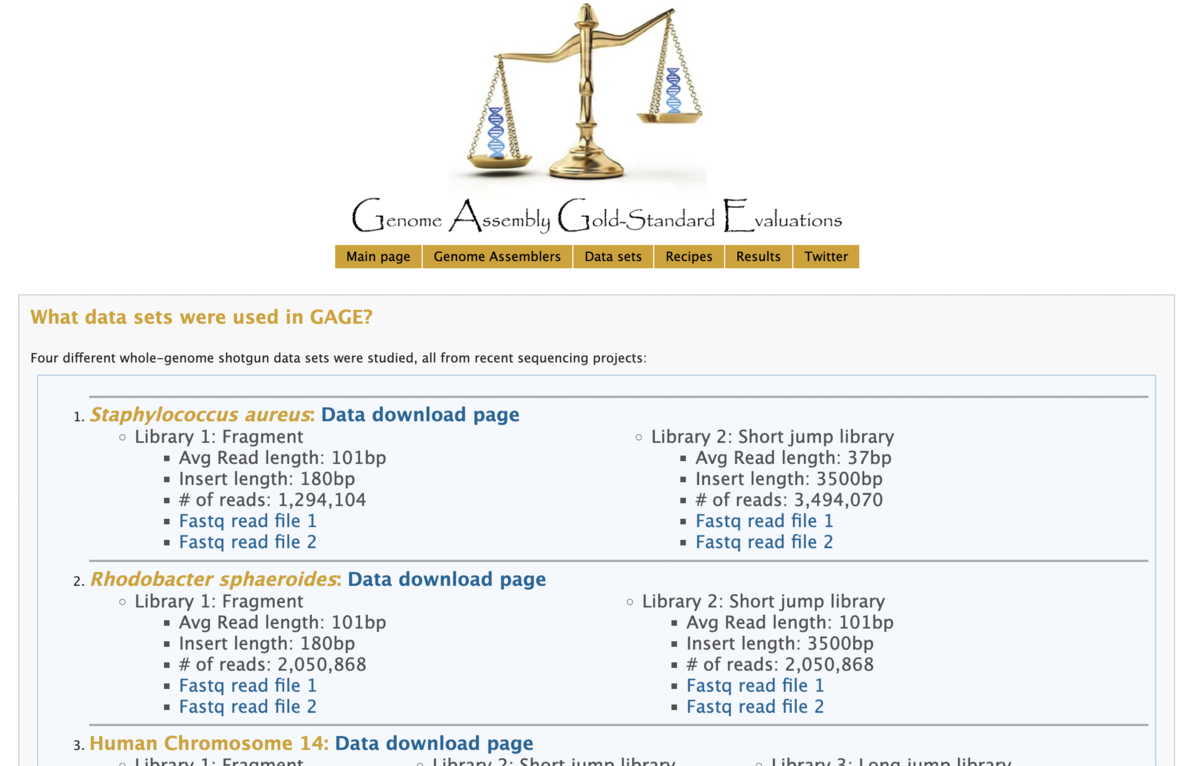
今回、GAGEから1. Staphylococcus aureus (黄色ブドウ球菌)のシーケンスデータをダウンロードします。
簡単なサンプル情報は下記に示します。
# テストデータ # Staphylococcus aureus: Data download page # Library 1: Fragment # Avg Read length: 101bp # Insert length: 180bp # # of reads: 1,294,104
まずは、Read1とRead2のFastqファイルをそれぞれwgetでダウンロードして、gzipで解凍します。
#ダウンロード wget http://gage.cbcb.umd.edu/data/Staphylococcus_aureus/Data.original/frag_1.fastq.gz http://gage.cbcb.umd.edu/data/Staphylococcus_aureus/Data.original/frag_2.fastq.gz #解凍 gzip -d -k frag_1.fastq.gz gzip -d -k frag_2.fastq.gz #ファイルの詳細表示 ls -lh #-rw-r--r-- 1 sas staff 140M 4 15 2011 frag_1.fastq #-rw-r--r-- 1 sas staff 61M 4 15 2011 frag_1.fastq.gz #-rw-r--r-- 1 sas staff 140M 4 15 2011 frag_2.fastq #-rw-r--r-- 1 sas staff 65M 4 15 2011 frag_2.fastq.gz
データは、fastq.gz形式で約60Mバイト、生データで約150MBバイトという、
低スペックPCにも優しい低容量です。
次に、実際に、flash2コマンドを使って、リードのマージを行います。
入力は、2つのfastqファイルを使います。
-tオプションでは、worker threadsの数を指定します。
#実行 flash2 frag_1.fastq frag_2.fastq -t 2 #[FLASH] Starting FLASH v2.2.00 #[FLASH] Fast Length Adjustment of SHort reads #[FLASH] #[FLASH] Input files: #[FLASH] frag_1.fastq #[FLASH] frag_2.fastq #[FLASH] #[FLASH] Output files: #[FLASH] ./out.extendedFrags.fastq #[FLASH] ./out.notCombined_1.fastq #[FLASH] ./out.notCombined_2.fastq #[FLASH] ./out.hist #[FLASH] ./out.histogram #[FLASH] #[FLASH] Parameters: #[FLASH] Min overlap: 10 #[FLASH] Min overlap outie: 35 #[FLASH] Max overlap: 65 #[FLASH] Max mismatch density: 0.250000 #[FLASH] Allow "outie" pairs: true #[FLASH] Cap mismatch quals: false #[FLASH] Combiner threads: 2 #[FLASH] Input format: FASTQ, phred_offset=33 #[FLASH] Output format: FASTQ, phred_offset=33 #[FLASH] #[FLASH] Starting reader and writer threads #[FLASH] Starting 2 combiner threads #[FLASH] Processed 25000 read pairs #[FLASH] Processed 50000 read pairs #[FLASH] Processed 75000 read pairs #[FLASH] Processed 100000 read pairs #[FLASH] Processed 125000 read pairs #[FLASH] Processed 150000 read pairs #[FLASH] Processed 175000 read pairs #[FLASH] Processed 200000 read pairs #[FLASH] Processed 225000 read pairs #[FLASH] Processed 250000 read pairs #[FLASH] Processed 275000 read pairs #[FLASH] Processed 300000 read pairs #[FLASH] Processed 325000 read pairs #[FLASH] Processed 350000 read pairs #[FLASH] Processed 375000 read pairs #[FLASH] Processed 400000 read pairs #[FLASH] Processed 425000 read pairs #[FLASH] Processed 450000 read pairs #[FLASH] Processed 475000 read pairs #[FLASH] Processed 500000 read pairs #[FLASH] Processed 525000 read pairs #[FLASH] Processed 550000 read pairs #[FLASH] Processed 575000 read pairs #[FLASH] Processed 600000 read pairs #[FLASH] Processed 625000 read pairs #[FLASH] Processed 647052 read pairs #[FLASH] #[FLASH] Read combination statistics: #[FLASH] Total pairs: 647052 #[FLASH] Discarded pairs: 262 #[FLASH] Percent Discarded: 0.04% #[FLASH] Combined pairs: 370303 #[FLASH] Innie pairs: 369148 (99.69% of combined) #[FLASH] Outie pairs: 1155 (0.31% of combined) #[FLASH] Uncombined pairs: 276487 #[FLASH] Percent combined: 57.25% #[FLASH] #[FLASH] Writing histogram files. #[FLASH] #[FLASH] FLASH v2.2.00 complete! #[FLASH] 11.136 seconds elapsed
解析が終って、出力結果(out.histogram)を出力表示すれば、テキストのヒストグラムとして結果を確認できます。 アスタリスクのヒストグラムは、なんとも乙な感じです。
#結果確認 #ヒストグラム確認 tail -n 90 out.histogram #103 #104 #105 #106 #107 #108 #109 #110 #111 #112 #113 #114 #115 * #116 * #117 * #118 * #119 * #120 * #121 ** #122 ** #123 *** #124 *** #125 **** #126 ***** #127 ****** #128 ******** #129 ******** #130 *********** #131 ************ #132 ************** #133 ***************** #134 ******************** #135 ********************** #136 ************************* #137 ***************************** #138 ******************************* #139 ************************************ #140 ************************************* #141 **************************************** #142 ********************************************* #143 ************************************************ #144 ************************************************** #145 ****************************************************** #146 ******************************************************** #147 *********************************************************** #148 ************************************************************* #149 *************************************************************** #150 **************************************************************** #151 ***************************************************************** #152 ******************************************************************* #153 ******************************************************************** #154 ********************************************************************** #155 ******************************************************************** #156 *********************************************************************** #157 ********************************************************************** #158 *********************************************************************** #159 ********************************************************************* #160 *********************************************************************** #161 ********************************************************************** #162 ************************************************************************ #163 *********************************************************************** #164 ********************************************************************* #165 ********************************************************************** #166 *********************************************************************** #167 ******************************************************************* #168 ******************************************************************* #169 ******************************************************************** #170 ****************************************************************** #171 ***************************************************************** #172 ***************************************************************** #173 **************************************************************** #174 **************************************************************** #175 ************************************************************* #176 ************************************************************* #177 ************************************************************** #178 ************************************************************** #179 ********************************************************** #180 ********************************************************** #181 ********************************************************** #182 ********************************************************* #183 ****************************************************** #184 ****************************************************** #185 ******************************************************* #186 ****************************************************** #187 ************************************************** #188 ************************************************** #189 **************************************************** #190 ****************************************************** #191 ********************************************** #192 ***********************************************
また、マージされたリードが出力されたFastq(out.extendedFrags.fastq)や ヒストグラムの数値のテキスト出力ファイルであるout.histもあります。
マージされなかったリードは、 out.notCombined_1.fastq、out.notCombined_2.fastqとしてアウトプットされます。
#Extended FASTQ 確認 head out.extendedFrags.fastq #@SRR022868.1483 #TCATACATATTAATATAGTCAGAACTAGTAATATAATTTTGGGCATTTCTATATAAATATCTATTCCATGACAGAAATACACATTGCGCTGGTCTTCCCATTTCTTTAAATAAATTTAAACGATTAATAATTGCTTTCTCT #+ #IIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIGIBECEIF8E;DEAE7<I1ICD2ICB%8I1354I25;A9E=>52?AI6;795:%6BIACII;II@<IIIIIA<0AFI3FIIIIIIIIIIIIIIIIIIIIII #@SRR022868.1630 #AATATGAAGCAATTTTTTCAAAAACGCTATTAGTCACAAAATGTAAACATAATTTTAACAAATGTGTGAACCCACGTCGCACCTTGATTTATCAACATTTTTGATACAATTTTATTAATTTTTTTCCCATAAGCCCT #+ #IIIIIIIIIIIIFIIIII>IIIBFIIIIII1II/5I:I5@6EI24%81+<,&+A4436(3,3,+*A+00(*#0/62,/&;&2.10-/3202+F+-8;+8,>31;?-:5%584+$)G2I6I438C9IIII.I,6)IIB #@SRR022868.1854 #TTCTGTGTATGAAAATATTTTTCATAAAAATTGTTTGAATCATGTAACAATCATATAAATTGCTGTTTATATTGTTGTGAAAATGAGTTGACAAAAGTCGGTATAGATATGTAGACATCCTATTTTTAGCGAG #ヒストグラムのヘッド表示 head out.hist #35 15 #36 12 #37 4 #38 11 #39 9 #40 7 #41 12 #42 10 #43 8 #44 11
まとめ
flash2を使って、ペアエンド・リードのマージをやってみました。 ソースからコンパイルすると、設定が少なくて助かります。 あと、M1 Macでもちゃんとコンパイルが通るもんなんですね。感心。
便利なバイオインフォのツールがたくさんありそうので、 引き続き、ツール発掘を続けてみます。
ゲノム解析の関連記事
参考資料
https://ccb.jhu.edu/software/FLASH/ccb.jhu.edu
補足
FASTQ形式からFASTA形式への変換 - sedコマンドを使う方法
sedコマンドを使って、Fastqファイルから必要な行を選択的に出力することができます。 4行ごとに1行目と2行目を出力すれば、配列のヘッダーとFASTAフォーマットに必要な配列が得られます。
下記のような実行例になります。
sed -n '1~4s/^@/>/p;2~4p' INFILE.fastq > OUTFILE.fasta
FASTQ形式からFASTA形式への変換 - fastq_to_fastaを使う方法
fastq_to_fastaは、FASTX-toolkitで利用可能です。巨大なデータセットでもうまく対応できます。下記のような実行例になります。
fastq_to_fasta -n -i INFILE.fastq -o OUTFILE.fasta
Anacondaの設定
結局使わなかったけども、、Anacondaの設定もメモしておきます
まずは、brewコマンドで、Anacondaをインストールします。
以下のbrew install --caskコマンドで入ります。
#インストール brew install --cask anaconda #... #==> Downloading https://repo.anaconda.com/archive/Anaconda3-2022.05-MacOSX-arm64 #==> Installing Cask anaconda #==> Running installer script 'Anaconda3-2022.05-MacOSX-arm64.sh' #パス設定 echo 'export PATH="/opt/homebrew/anaconda3/bin:$PATH"' >> .zshrc #tail -n 1 .zshrc source .zshrc #パス確認 echo $PATH #condaパス確認 which conda #/opt/homebrew/anaconda3/bin/conda
FLASH-1.2.11のインストール方法 from sourceforge.net
これは、FLASHのオリジナル版のインストール方法です。
ターミナルを起動して、以下のコマンドを実行していきます。
#ダウンロード wget https://sourceforge.net/projects/flashpage/files/FLASH-1.2.11.tar.gz #解凍 tar xzf FLASH-1.2.11.tar.gz #ディレクトリ移動 cd FLASH-1.2.11 #コンパイル make #パス設定の前段階: 手っ取り早く"/opt/homebrew/bin"に置いておく cd ../ mv FLASH-1.2.11 /opt/homebrew/bin #HOMEに移動 cd ~/ #パスを.zshrcを書き込む echo 'export PATH="/opt/homebrew/bin/FLASH-1.2.11:$PATH"' >> .zshrc #いったんソースする source .zshrc #パス設定 which flash #ヘルプで動作確認: 出力表示は補足に記載 flash -h
flash -h の出力
Usage: flash [OPTIONS] MATES_1.FASTQ MATES_2.FASTQ
flash [OPTIONS] --interleaved-input (MATES.FASTQ | -)
flash [OPTIONS] --tab-delimited-input (MATES.TAB | -)
----------------------------------------------------------------------------
DESCRIPTION
----------------------------------------------------------------------------
FLASH (Fast Length Adjustment of SHort reads) is an accurate and fast tool
to merge paired-end reads that were generated from DNA fragments whose
lengths are shorter than twice the length of reads. Merged read pairs result
in unpaired longer reads, which are generally more desired in genome
assembly and genome analysis processes.
Briefly, the FLASH algorithm considers all possible overlaps at or above a
minimum length between the reads in a pair and chooses the overlap that
results in the lowest mismatch density (proportion of mismatched bases in
the overlapped region). Ties between multiple overlaps are broken by
considering quality scores at mismatch sites. When building the merged
sequence, FLASH computes a consensus sequence in the overlapped region.
More details can be found in the original publication
(http://bioinformatics.oxfordjournals.org/content/27/21/2957.full).
Limitations of FLASH include:
- FLASH cannot merge paired-end reads that do not overlap.
- FLASH is not designed for data that has a significant amount of indel
errors (such as Sanger sequencing data). It is best suited for Illumina
data.
----------------------------------------------------------------------------
MANDATORY INPUT
----------------------------------------------------------------------------
The most common input to FLASH is two FASTQ files containing read 1 and read 2
of each mate pair, respectively, in the same order.
Alternatively, you may provide one FASTQ file, which may be standard input,
containing paired-end reads in either interleaved FASTQ (see the
--interleaved-input option) or tab-delimited (see the --tab-delimited-input
option) format. In all cases, gzip compressed input is autodetected. Also,
in all cases, the PHRED offset is, by default, assumed to be 33; use the
--phred-offset option to change it.
----------------------------------------------------------------------------
OUTPUT
----------------------------------------------------------------------------
The default output of FLASH consists of the following files:
- out.extendedFrags.fastq The merged reads.
- out.notCombined_1.fastq Read 1 of mate pairs that were not merged.
- out.notCombined_2.fastq Read 2 of mate pairs that were not merged.
- out.hist Numeric histogram of merged read lengths.
- out.histogram Visual histogram of merged read lengths.
FLASH also logs informational messages to standard output. These can also be
redirected to a file, as in the following example:
$ flash reads_1.fq reads_2.fq 2>&1 | tee flash.log
In addition, FLASH supports several features affecting the output:
- Writing the merged reads directly to standard output (--to-stdout)
- Writing gzip compressed output files (-z) or using an external
compression program (--compress-prog)
- Writing the uncombined read pairs in interleaved FASTQ format
(--interleaved-output)
- Writing all output reads to a single file in tab-delimited format
(--tab-delimited-output)
----------------------------------------------------------------------------
OPTIONS
----------------------------------------------------------------------------
-m, --min-overlap=NUM The minimum required overlap length between two
reads to provide a confident overlap. Default:
10bp.
-M, --max-overlap=NUM Maximum overlap length expected in approximately
90% of read pairs. It is by default set to 65bp,
which works well for 100bp reads generated from a
180bp library, assuming a normal distribution of
fragment lengths. Overlaps longer than the maximum
overlap parameter are still considered as good
overlaps, but the mismatch density (explained below)
is calculated over the first max_overlap bases in
the overlapped region rather than the entire
overlap. Default: 65bp, or calculated from the
specified read length, fragment length, and fragment
length standard deviation.
-x, --max-mismatch-density=NUM
Maximum allowed ratio between the number of
mismatched base pairs and the overlap length.
Two reads will not be combined with a given overlap
if that overlap results in a mismatched base density
higher than this value. Note: Any occurence of an
'N' in either read is ignored and not counted
towards the mismatches or overlap length. Our
experimental results suggest that higher values of
the maximum mismatch density yield larger
numbers of correctly merged read pairs but at
the expense of higher numbers of incorrectly
merged read pairs. Default: 0.25.
-O, --allow-outies Also try combining read pairs in the "outie"
orientation, e.g.
Read 1: <-----------
Read 2: ------------>
as opposed to only the "innie" orientation, e.g.
Read 1: <------------
Read 2: ----------->
FLASH uses the same parameters when trying each
orientation. If a read pair can be combined in
both "innie" and "outie" orientations, the
better-fitting one will be chosen using the same
scoring algorithm that FLASH normally uses.
This option also causes extra .innie and .outie
histogram files to be produced.
-p, --phred-offset=OFFSET
The smallest ASCII value of the characters used to
represent quality values of bases in FASTQ files.
It should be set to either 33, which corresponds
to the later Illumina platforms and Sanger
platforms, or 64, which corresponds to the
earlier Illumina platforms. Default: 33.
-r, --read-len=LEN
-f, --fragment-len=LEN
-s, --fragment-len-stddev=LEN
Average read length, fragment length, and fragment
standard deviation. These are convenience parameters
only, as they are only used for calculating the
maximum overlap (--max-overlap) parameter.
The maximum overlap is calculated as the overlap of
average-length reads from an average-size fragment
plus 2.5 times the fragment length standard
deviation. The default values are -r 100, -f 180,
and -s 18, so this works out to a maximum overlap of
65 bp. If --max-overlap is specified, then the
specified value overrides the calculated value.
If you do not know the standard deviation of the
fragment library, you can probably assume that the
standard deviation is 10% of the average fragment
length.
--cap-mismatch-quals Cap quality scores assigned at mismatch locations
to 2. This was the default behavior in FLASH v1.2.7
and earlier. Later versions will instead calculate
such scores as max(|q1 - q2|, 2); that is, the
absolute value of the difference in quality scores,
but at least 2. Essentially, the new behavior
prevents a low quality base call that is likely a
sequencing error from significantly bringing down
the quality of a high quality, likely correct base
call.
--interleaved-input Instead of requiring files MATES_1.FASTQ and
MATES_2.FASTQ, allow a single file MATES.FASTQ that
has the paired-end reads interleaved. Specify "-"
to read from standard input.
--interleaved-output Write the uncombined pairs in interleaved FASTQ
format.
-I, --interleaved Equivalent to specifying both --interleaved-input
and --interleaved-output.
-Ti, --tab-delimited-input
Assume the input is in tab-delimited format
rather than FASTQ, in the format described below in
'--tab-delimited-output'. In this mode you should
provide a single input file, each line of which must
contain either a read pair (5 fields) or a single
read (3 fields). FLASH will try to combine the read
pairs. Single reads will be written to the output
file as-is if also using --tab-delimited-output;
otherwise they will be ignored. Note that you may
specify "-" as the input file to read the
tab-delimited data from standard input.
-To, --tab-delimited-output
Write output in tab-delimited format (not FASTQ).
Each line will contain either a combined pair in the
format 'tag <tab> seq <tab> qual' or an uncombined
pair in the format 'tag <tab> seq_1 <tab> qual_1
<tab> seq_2 <tab> qual_2'.
-o, --output-prefix=PREFIX
Prefix of output files. Default: "out".
-d, --output-directory=DIR
Path to directory for output files. Default:
current working directory.
-c, --to-stdout Write the combined reads to standard output. In
this mode, with FASTQ output (the default) the
uncombined reads are discarded. With tab-delimited
output, uncombined reads are included in the
tab-delimited data written to standard output.
In both cases, histogram files are not written,
and informational messages are sent to standard
error rather than to standard output.
-z, --compress Compress the output files directly with zlib,
using the gzip container format. Similar to
specifying --compress-prog=gzip and --suffix=gz,
but may be slightly faster.
--compress-prog=PROG Pipe the output through the compression program
PROG, which will be called as `PROG -c -',
plus any arguments specified by --compress-prog-args.
PROG must read uncompressed data from standard input
and write compressed data to standard output when
invoked as noted above.
Examples: gzip, bzip2, xz, pigz.
--compress-prog-args=ARGS
A string of additional arguments that will be passed
to the compression program if one is specified with
--compress-prog=PROG. (The arguments '-c -' are
still passed in addition to explicitly specified
arguments.)
--suffix=SUFFIX, --output-suffix=SUFFIX
Use SUFFIX as the suffix of the output files
after ".fastq". A dot before the suffix is assumed,
unless an empty suffix is provided. Default:
nothing; or 'gz' if -z is specified; or PROG if
--compress-prog=PROG is specified.
-t, --threads=NTHREADS Set the number of worker threads. This is in
addition to the I/O threads. Default: number of
processors. Note: if you need FLASH's output to
appear deterministically or in the same order as
the original reads, you must specify -t 1
(--threads=1).
-q, --quiet Do not print informational messages.
-h, --help Display this help and exit.
-v, --version Display version.
Run `flash --help | less' to prevent this text from scrolling by.
flash2 --h の出力
Usage: flash [OPTIONS] MATES_1.FASTQ MATES_2.FASTQ
flash [OPTIONS] --interleaved-input (MATES.FASTQ | -)
flash [OPTIONS] --tab-delimited-input (MATES.TAB | -)
----------------------------------------------------------------------------
DESCRIPTION
----------------------------------------------------------------------------
FLASH (Fast Length Adjustment of SHort reads) is an accurate and fast tool
to merge paired-end reads that were generated from DNA fragments whose
lengths are shorter than twice the length of reads. Merged read pairs result
in unpaired longer reads, which are generally more desired in genome
assembly and genome analysis processes.
Briefly, the FLASH algorithm considers all possible overlaps at or above a
minimum length between the reads in a pair and chooses the overlap that
results in the lowest mismatch density (proportion of mismatched bases in
the overlapped region). Ties between multiple overlaps are broken by
considering quality scores at mismatch sites. When building the merged
sequence, FLASH computes a consensus sequence in the overlapped region.
More details can be found in the original publication
(http://bioinformatics.oxfordjournals.org/content/27/21/2957.full).
Limitations of FLASH include:
- FLASH cannot merge paired-end reads that do not overlap.
- FLASH is not designed for data that has a significant amount of indel
errors (such as Sanger sequencing data). It is best suited for Illumina
data.
----------------------------------------------------------------------------
MANDATORY INPUT
----------------------------------------------------------------------------
The most common input to FLASH is two FASTQ files containing read 1 and read 2
of each mate pair, respectively, in the same order.
Alternatively, you may provide one FASTQ file, which may be standard input,
containing paired-end reads in either interleaved FASTQ (see the
--interleaved-input option) or tab-delimited (see the --tab-delimited-input
option) format. In all cases, gzip compressed input is autodetected. Also,
in all cases, the PHRED offset is, by default, assumed to be 33; use the
--phred-offset option to change it.
----------------------------------------------------------------------------
OUTPUT
----------------------------------------------------------------------------
The default output of FLASH consists of the following files:
- out.extendedFrags.fastq The merged reads.
- out.notCombined_1.fastq Read 1 of mate pairs that were not merged.
- out.notCombined_2.fastq Read 2 of mate pairs that were not merged.
- out.hist Numeric histogram of merged read lengths.
- out.histogram Visual histogram of merged read lengths.
FLASH also logs informational messages to standard output. These can also be
redirected to a file, as in the following example:
$ flash reads_1.fq reads_2.fq 2>&1 | tee flash.log
In addition, FLASH supports several features affecting the output:
- Writing the merged reads directly to standard output (--to-stdout)
- Writing gzip compressed output files (-z) or using an external
compression program (--compress-prog)
- Writing the uncombined read pairs in interleaved FASTQ format
(--interleaved-output)
- Writing all output reads to a single file in tab-delimited format
(--tab-delimited-output)
----------------------------------------------------------------------------
OPTIONS
----------------------------------------------------------------------------
-Q, --quality-cutoff=NUM The cut off number for the quality score
corresponding wtih the percent cutoff. Default:
2.
-C, --percent-cutoff=NUM The cutoff percentage for each read that will
be discarded if it falls below -Q option. (0-100) Default:
50.
-D, --no-discard This turns off the discard logic Default: false
-m, --min-overlap=NUM The minimum required overlap length between two
reads to provide a confident overlap. Default 10bp.
-M, --max-overlap=NUM Maximum overlap length expected in approximately
90% of read pairs. It is by default set to 65bp,
which works well for 100bp reads generated from a
180bp library, assuming a normal distribution of
fragment lengths. Overlaps longer than the maximum
overlap parameter are still considered as good
overlaps, but the mismatch density (explained below)
is calculated over the first max_overlap bases in
the overlapped region rather than the entire
overlap. Default: 65bp, or calculated from the
specified read length, fragment length, and fragment
length standard deviation.
-e, --min-overlap-outie=NUM The minimum required overlap length between two
reads to provide a confident overlap in an outie scenario.
Default: 35bp.
-x, --max-mismatch-density=NUM
Maximum allowed ratio between the number of
mismatched base pairs and the overlap length.
Two reads will not be combined with a given overlap
if that overlap results in a mismatched base density
higher than this value. Note: Any occurence of an
'N' in either read is ignored and not counted
towards the mismatches or overlap length. Our
experimental results suggest that higher values of
the maximum mismatch density yield larger
numbers of correctly merged read pairs but at
the expense of higher numbers of incorrectly
merged read pairs. Default: 0.25.
-O, --allow-outies Also try combining read pairs in the "outie"
orientation, e.g.
Read 1: <-----------
Read 2: ------------>
as opposed to only the "innie" orientation, e.g.
Read 1: <------------
Read 2: ----------->
FLASH uses the same parameters when trying each
orientation. If a read pair can be combined in
both "innie" and "outie" orientations, the
better-fitting one will be chosen using the same
scoring algorithm that FLASH normally uses.
This option also causes extra .innie and .outie
histogram files to be produced.
-p, --phred-offset=OFFSET
The smallest ASCII value of the characters used to
represent quality values of bases in FASTQ files.
It should be set to either 33, which corresponds
to the later Illumina platforms and Sanger
platforms, or 64, which corresponds to the
earlier Illumina platforms. Default: 33.
-r, --read-len=LEN
-f, --fragment-len=LEN
-s, --fragment-len-stddev=LEN
Average read length, fragment length, and fragment
standard deviation. These are convenience parameters
only, as they are only used for calculating the
maximum overlap (--max-overlap) parameter.
The maximum overlap is calculated as the overlap of
average-length reads from an average-size fragment
plus 2.5 times the fragment length standard
deviation. The default values are -r 100, -f 180,
and -s 18, so this works out to a maximum overlap of
65 bp. If --max-overlap is specified, then the
specified value overrides the calculated value.
If you do not know the standard deviation of the
fragment library, you can probably assume that the
standard deviation is 10% of the average fragment
length.
--cap-mismatch-quals Cap quality scores assigned at mismatch locations
to 2. This was the default behavior in FLASH v1.2.7
and earlier. Later versions will instead calculate
such scores as max(|q1 - q2|, 2); that is, the
absolute value of the difference in quality scores,
but at least 2. Essentially, the new behavior
prevents a low quality base call that is likely a
sequencing error from significantly bringing down
the quality of a high quality, likely correct base
call.
--interleaved-input Instead of requiring files MATES_1.FASTQ and
MATES_2.FASTQ, allow a single file MATES.FASTQ that
has the paired-end reads interleaved. Specify "-"
to read from standard input.
--interleaved-output Write the uncombined pairs in interleaved FASTQ
format.
-I, --interleaved Equivalent to specifying both --interleaved-input
and --interleaved-output.
-Ti, --tab-delimited-input
Assume the input is in tab-delimited format
rather than FASTQ, in the format described below in
'--tab-delimited-output'. In this mode you should
provide a single input file, each line of which must
contain either a read pair (5 fields) or a single
read (3 fields). FLASH will try to combine the read
pairs. Single reads will be written to the output
file as-is if also using --tab-delimited-output;
otherwise they will be ignored. Note that you may
specify "-" as the input file to read the
tab-delimited data from standard input.
-To, --tab-delimited-output
Write output in tab-delimited format (not FASTQ).
Each line will contain either a combined pair in the
format 'tag <tab> seq <tab> qual' or an uncombined
pair in the format 'tag <tab> seq_1 <tab> qual_1
<tab> seq_2 <tab> qual_2'.
-o, --output-prefix=PREFIX
Prefix of output files. Default: "out".
-d, --output-directory=DIR
Path to directory for output files. Default:
current working directory.
-c, --to-stdout Write the combined reads to standard output. In
this mode, with FASTQ output (the default) the
uncombined reads are discarded. With tab-delimited
output, uncombined reads are included in the
tab-delimited data written to standard output.
In both cases, histogram files are not written,
and informational messages are sent to standard
error rather than to standard output.
-z, --compress Compress the output files directly with zlib,
using the gzip container format. Similar to
specifying --compress-prog=gzip and --suffix=gz,
but may be slightly faster.
--compress-prog=PROG Pipe the output through the compression program
PROG, which will be called as `PROG -c -',
plus any arguments specified by --compress-prog-args.
PROG must read uncompressed data from standard input
and write compressed data to standard output when
invoked as noted above.
Examples: gzip, bzip2, xz, pigz.
--compress-prog-args=ARGS
A string of additional arguments that will be passed
to the compression program if one is specified with
--compress-prog=PROG. (The arguments '-c -' are
still passed in addition to explicitly specified
arguments.)
--suffix=SUFFIX, --output-suffix=SUFFIX
Use SUFFIX as the suffix of the output files
after ".fastq". A dot before the suffix is assumed,
unless an empty suffix is provided. Default:
nothing; or 'gz' if -z is specified; or PROG if
--compress-prog=PROG is specified.
-t, --threads=NTHREADS Set the number of worker threads. This is in
addition to the I/O threads. Default: number of
processors. Note: if you need FLASH's output to
appear deterministically or in the same order as
the original reads, you must specify -t 1
(--threads=1).
-q, --quiet Do not print informational messages.
-h, --help Display this help and exit.
-v, --version Display version.
Run `flash2 --help | less' to prevent this text from scrolling by.